Across many scientific disciplines, but in particular in the field of genomics, major breakthroughs have often resulted from new technologies. From Sanger sequencing, which made it possible to sequence the human genome, to the microarray technologies that enabled the first large-scale genome-wide experiments, new instruments and tools have allowed us to look ever more deeply into the genome and apply the results broadly to health, agriculture and ecology.
One of the most transformative new technologies in genomics was high-throughput sequencing (HTS), which first became commercially available in the early 2000s. HTS allowed scientists and clinicians to produce sequencing data quickly, cheaply, and at scale. However, the output of HTS instruments is not the genome sequence for the individual being analyzed — for humans this is 3 billion paired bases (guanine, cytosine, adenine and thymine) organized into 23 pairs of chromosomes. Instead, these instruments generate ~1 billion short sequences, known as reads. Each read represents just 100 of the 3 billion bases, and per-base error rates range from 0.1-10%. Processing the HTS output into a single, accurate and complete genome sequence is a major outstanding challenge. The importance of this problem, for biomedical applications in particular, has motivated efforts such as the Genome in a Bottle Consortium (GIAB), which produces high confidence human reference genomes that can be used for validation and benchmarking, as well as the precisionFDA community challenges, which are designed to foster innovation that will improve the quality and accuracy of HTS-based genomic tests.
CAPTION: For any given location in the genome, there are multiple reads among the ~1 billion that include a base at that position. Each read is aligned to a reference, and then each of the bases in the read is compared to the base of the reference at that location. When a read includes a base that differs from the reference, it may indicate a variant (a difference in the true sequence), or it may be an error.
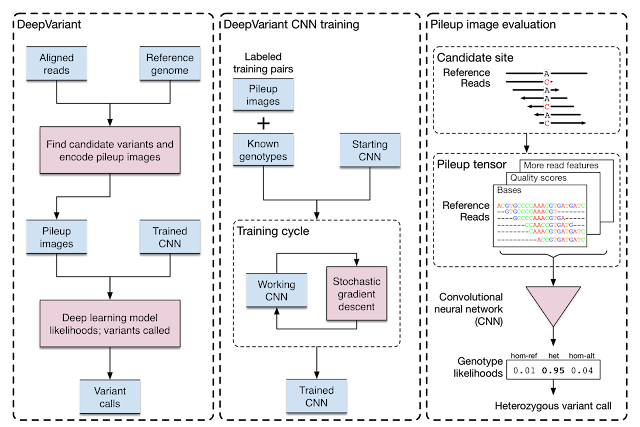
Today, we announce the open source release of DeepVariant, a deep learning technology to reconstruct the true genome sequence from HTS sequencer data with significantly greater accuracy than previous classical methods. This work is the product of more than two years of research by the Google Brain team, in collaboration with Verily Life Sciences. DeepVariant transforms the task of variant calling, as this reconstruction problem is known in genomics, into an image classification problem well-suited to Google's existing technology and expertise.
CAPTION: Each of the four images above is a visualization of actual sequencer reads aligned to a reference genome. A key question is how to use the reads to determine whether there is a variant on both chromosomes, on just one chromosome, or on neither chromosome. There is more than one type of variant, with SNPs and insertions/deletions being the most common. A: a true SNP on one chromosome pair, B: a deletion on one chromosome, C: a deletion on both chromosomes, D: a false variant caused by errors. It's easy to see that these look quite distinct when visualized in this manner.
We started with GIAB reference genomes, for which there is high-quality ground truth (or the closest approximation currently possible). Using multiple replicates of these genomes, we produced tens of millions of training examples in the form of multi-channel tensors encoding the HTS instrument data, and then trained a TensorFlow-based image classification model to identify the true genome sequence from the experimental data produced by the instruments. Although the resulting deep learning model, DeepVariant, had no specialized knowledge about genomics or HTS, within a year it had won the the highest SNP accuracy award at the precisionFDA Truth Challenge, outperforming state-of-the-art methods. Since then, we've further reduced the error rate by more than 50%.
DeepVariant is being released as open source software to encourage collaboration and to accelerate the use of this technology to solve real world problems. To further this goal, we partnered with Google Cloud Platform (GCP) to deploy DeepVariant workflows on GCP, available today, in configurations optimized for low-cost and fast turnarounds using scalable GCP technologies like the Pipelines API. This paired set of releases provides a smooth ramp for users to explore and evaluate the capabilities of DeepVariant in their current compute environment while providing a scalable, cloud-based solution to satisfy the needs of even the largest genomics datasets.
DeepVariant is the first of what we hope will be many contributions that leverage Google's computing infrastructure and ML expertise to both better understand the genome and to provide deep learning-based genomics tools to the community. This is all part of a broader goal to apply Google technologies to healthcare and other scientific applications, and to make the results of these efforts broadly accessible.
By Mark DePristo and Ryan Poplin, Google Brain Team