Posted by the TensorFlow TeamWelcome to Part 3 of a blog series that introduces TensorFlow Datasets and Estimators. Part 1 focused on pre-made Estimators, while Part 2 discussed feature columns. Here in Part 3, you'll learn how to create your own custom Estimators. In particular, we're going to demonstrate how to create a custom Estimator that mimics DNNClassifier's behavior when solving the Iris problem.
If you are feeling impatient, feel free to compare and contrast the following full programs:
- Source code for Iris implemented with the pre-made DNNClassifier Estimator here.
- Source code for Iris implemented with the custom Estimator here.
Pre-made vs. custom
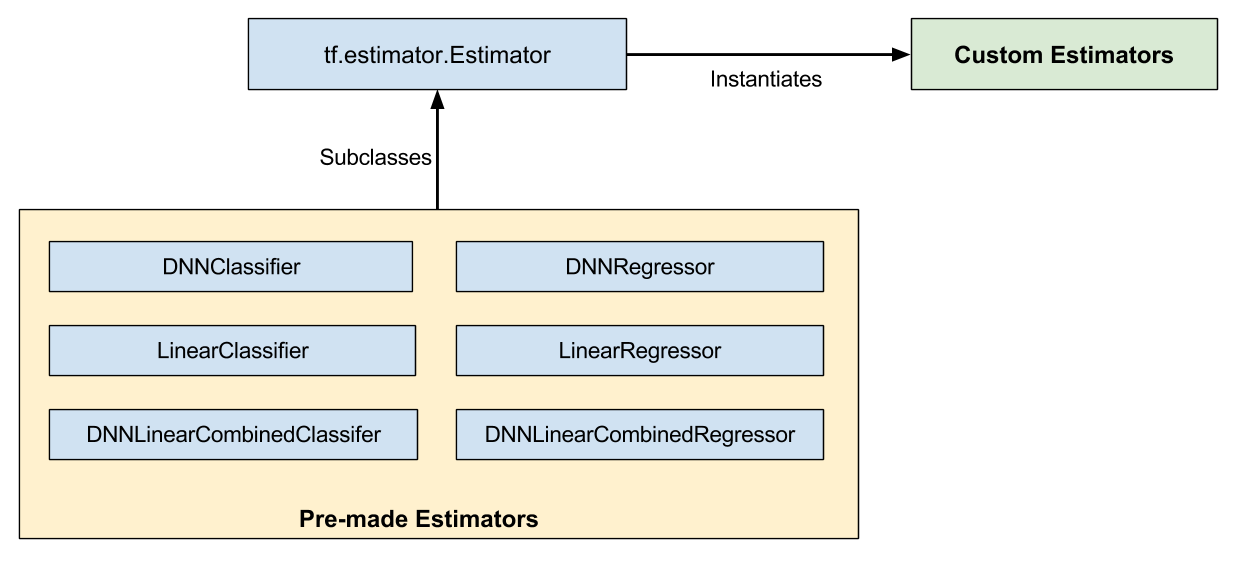
As Figure 1 shows, pre-made Estimators are subclasses of the tf.estimator.Estimatorbase class, while custom Estimators are an instantiation of tf.estimator.Estimator:
Figure 1. Pre-made and custom Estimators are all Estimators.
Pre-made Estimators are fully-baked. Sometimes though, you need more control over an Estimator's behavior. That's where custom Estimators come in.
You can create a custom Estimator to do just about anything. If you want hidden layers connected in some unusual fashion, write a custom Estimator. If you want to calculate a unique metric for your model, write a custom Estimator. Basically, if you want an Estimator optimized for your specific problem, write a custom Estimator.
A model function (model_fn) implements your model. The only difference between working with pre-made Estimators and custom Estimators is:
- With pre-made Estimators, someone already wrote the model function for you.
- With custom Estimators, you must write the model function.
Your model function could implement a wide range of algorithms, defining all sorts of hidden layers and metrics. Like input functions, all model functions must accept a standard group of input parameters and return a standard group of output values. Just as input functions can leverage the Dataset API, model functions can leverage the Layers API and the Metrics API.
Iris as a pre-made Estimator: A quick refresher
Before demonstrating how to implement Iris as a custom Estimator, we wanted to remind you how we implemented Iris as a pre-made Estimator in Part 1 of this series. In that Part, we created a fully connected, deepneural network for the Iris dataset simply by instantiating a pre-made Estimator as follows:
# Instantiate a deep neural network classifier.
classifier = tf.estimator.DNNClassifier(
feature_columns=feature_columns, # The input features to our model.
hidden_units=[10, 10], # Two layers, each with 10 neurons.
n_classes=3, # The number of output classes (three Iris species).
model_dir=PATH) # Pathname of directory where checkpoints, etc. are stored.
The preceding code creates a deep neural network with the following characteristics:
- A list of feature columns. (The definitions of the feature columns are not shown in the preceding snippet.) For Iris, the feature columns are numeric representations of four input features.
- Two fully connected layers, each having 10 neurons. A fully connected layer (also called a dense layer) is connected to every neuron in the subsequent layer.
- An output layer consisting of a three-element list. The elements of that list are all floating-point values; the sum of those values must be 1.0 (this is a probability distribution).
- A directory (
PATH) in which the trained model and various checkpoints will be stored.
Figure 2 illustrates the input layer, hidden layers, and output layer of the Iris model. For reasons pertaining to clarity, we've only drawn 4 of the nodes in each hidden layer.
Figure 2. Our implementation of Iris contains four features, two hidden layers, and a logits output layer.
Let's see how to solve the same Iris problem with a custom Estimator.
Input function
One of the biggest advantages of the Estimator framework is that you can experiment with different algorithms without changing your data pipeline. We will therefore reuse much of the input function from Part 1:
def my_input_fn(file_path, repeat_count=1, shuffle_count=1):
def decode_csv(line):
parsed_line = tf.decode_csv(line, [[0.], [0.], [0.], [0.], [0]])
label = parsed_line[-1] # Last element is the label
del parsed_line[-1] # Delete last element
features = parsed_line # Everything but last elements are the features
d = dict(zip(feature_names, features)), label
return d
dataset = (tf.data.TextLineDataset(file_path) # Read text file
.skip(1) # Skip header row
.map(decode_csv, num_parallel_calls=4) # Decode each line
.cache() # Warning: Caches entire dataset, can cause out of memory
.shuffle(shuffle_count) # Randomize elems (1 == no operation)
.repeat(repeat_count) # Repeats dataset this # times
.batch(32)
.prefetch(1) # Make sure you always have 1 batch ready to serve
)
iterator = dataset.make_one_shot_iterator()
batch_features, batch_labels = iterator.get_next()
return batch_features, batch_labels
Notice that the input function returns the following two values:
batch_features, which is a dictionary. The dictionary's keys are the names of the features, and the dictionary's values are the feature's values. batch_labels, which is a list of the label's values for a batch.
Refer to Part 1 for full details on input functions.
Create feature columns
As detailed in Part 2 of our series, you must define your model's feature columns to specify the representation of each feature. Whether working with pre-made Estimators or custom Estimators, you define feature columns in the same fashion. For example, the following code creates feature columns representing the four features (all numerical) in the Iris dataset:
feature_columns = [
tf.feature_column.numeric_column(feature_names[0]),
tf.feature_column.numeric_column(feature_names[1]),
tf.feature_column.numeric_column(feature_names[2]),
tf.feature_column.numeric_column(feature_names[3])
]
Write a model function
We are now ready to write the model_fn for our custom Estimator. Let's start with the function declaration:
def my_model_fn(
features, # This is batch_features from input_fn
labels, # This is batch_labels from input_fn
mode): # Instance of tf.estimator.ModeKeys, see below
The first two arguments are the features and labels returned from the input function; that is, features and labels are the handles to the data your model will use. The mode argument indicates whether the caller is requesting training, predicting, or evaluating.
To implement a typical model function, you must do the following:
- Define the model's layers.
- Specify the model's behavior in three the different modes.
Define the model's layers
If your custom Estimator generates a deep neural network, you must define the following three layers:
- an input layer
- one or more hidden layers
- an output layer
Use the Layers API (tf.layers) to define hidden and output layers.
If your custom Estimator generates a linear model, then you only have to generate a single layer, which we'll describe in the next section.
Define the input layer
Call tf.feature_column.input_layerto define the input layer for a deep neural network. For example:
# Create the layer of input
input_layer = tf.feature_column.input_layer(features, feature_columns)
The preceding line creates our input layer, reading our featuresthrough the input function and filtering them through the feature_columns defined earlier. See Part 2 for details on various ways to represent data through feature columns.
To create the input layer for a linear model, call tf.feature_column.linear_modelinstead of tf.feature_column.input_layer. Since a linear model has no hidden layers, the returned value from tf.feature_column.linear_model serves as both the input layer and output layer. In other words, the returned value from this function isthe prediction.
Establish Hidden Layers
If you are creating a deep neural network, you must define one or more hidden layers. The Layers API provides a rich set of functions to define all types of hidden layers, including convolutional, pooling, and dropout layers. For Iris, we're simply going to call tf.layers.Densetwice to create two dense hidden layers, each with 10 neurons. By "dense," we mean that each neuron in the first hidden layer is connected to each neuron in the second hidden layer. Here's the relevant code:
# Definition of hidden layer: h1
# (Dense returns a Callable so we can provide input_layer as argument to it)
h1 = tf.layers.Dense(10, activation=tf.nn.relu)(input_layer)
# Definition of hidden layer: h2
# (Dense returns a Callable so we can provide h1 as argument to it)
h2 = tf.layers.Dense(10, activation=tf.nn.relu)(h1)
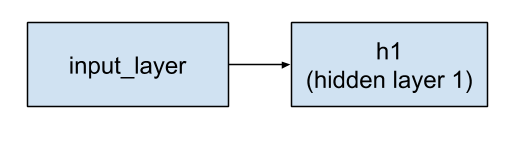
The inputs parameter to tf.layers.Dense identifies the preceding layer. The layer preceding h1 is the input layer.
Figure 3. The input layer feeds into hidden layer 1.
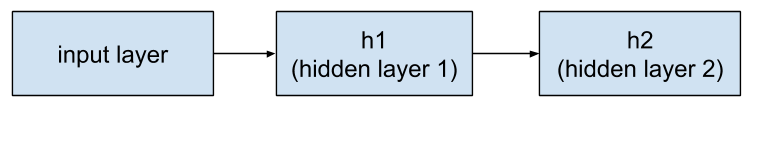
The preceding layer to h2 is h1. So, the string of layers now looks like this:
Figure 4. Hidden layer 1 feeds into hidden layer 2.
The first argument to tf.layers.Densedefines the number of its output neurons—10 in this case.
The activation parameter defines the activation function—Relu in this case.
Note that tf.layers.Denseprovides many additional capabilities, including the ability to set a multitude of regularization parameters. For the sake of simplicity, though, we're going to simply accept the default values of the other parameters. Also, when looking at tf.layersyou may encounter lower-case versions (e.g. tf.layers.dense). As a general rule, you should use the class versions which start with a capital letter (tf.layers.Dense).
Output Layer
We'll define the output layer by calling tf.layers.Dense yet again:
# Output 'logits' layer is three numbers = probability distribution
# (Dense returns a Callable so we can provide h2 as argument to it)
logits = tf.layers.Dense(3)(h2)
Notice that the output layer receives its input from h2. Therefore, the full set of layers is now connected as follows:
Figure 5. Hidden layer 2 feeds into the output layer.
When defining an output layer, the units parameter specifies the number of possible output values. So, by setting units to 3, the tf.layers.Densefunction establishes a three-element logits vector. Each cell of the logits vector contains the probability of the Iris being Setosa, Versicolor, or Virginica, respectively.
Since the output layer is a final layer, the call to tf.layers.Denseomits the optional activation parameter.
Implement training, evaluation, and prediction
The final step in creating a model function is to write branching code that implements prediction, evaluation, and training.
The model function gets invoked whenever someone calls the Estimator's train, evaluate, or predict methods. Recall that the signature for the model function looks like this:
def my_model_fn(
features, # This is batch_features from input_fn
labels, # This is batch_labels from input_fn
mode): # Instance of tf.estimator.ModeKeys, see below
Focus on that third argument, mode. As the following table shows, when someone calls train, evaluate, or predict, the Estimator framework invokes your model function with the mode parameter set as follows:
Table 2. Values of mode.
| Caller invokes custom Estimator method... | Estimator framework calls your model function with the mode parameter set to... |
train() | ModeKeys.TRAIN |
evaluate() | ModeKeys.EVAL |
predict() | ModeKeys.PREDICT |
For example, suppose you instantiate a custom Estimator to generate an object named classifier. Then, you might make the following call (never mind the parameters to my_input_fn at this time):
classifier.train(
input_fn=lambda: my_input_fn(FILE_TRAIN, repeat_count=500, shuffle_count=256))
The Estimator framework then calls your model function with mode set to ModeKeys.TRAIN.
Your model function must provide code to handle all three of the mode values. For each mode value, your code must return an instance of tf.estimator.EstimatorSpec, which contains the information the caller requires. Let's examine each mode.
PREDICT
When model_fn is called with mode == ModeKeys.PREDICT, the model function must return a tf.estimator.EstimatorSpeccontaining the following information:
- the mode, which is
tf.estimator.ModeKeys.PREDICT - the prediction
The model must have been trained prior to making a prediction. The trained model is stored on disk in the directory established when you instantiated the Estimator.
For our case, the code to generate the prediction looks as follows:
# class_ids will be the model prediction for the class (Iris flower type)
# The output node with the highest value is our prediction
predictions = { 'class_ids': tf.argmax(input=logits, axis=1) }
# Return our prediction
if mode == tf.estimator.ModeKeys.PREDICT:
return tf.estimator.EstimatorSpec(mode, predictions=predictions)
The block is surprisingly brief--the lines of code are simply the bucket at the end of a long hose that catches the falling predictions. After all, the Estimator has already done all the heavy lifting to make a prediction:
- The input function provides the model function with data (feature values) to infer from.
- The model function transforms those feature values into feature columns.
- The model function runs those feature columns through the previously-trained model.
The output layer is a logits vector that contains the value of each of the three Iris species being the input flower. The tf.argmaxmethod selects the Iris species in that logits vector with the highest value.
Notice that the highest value is assigned to a dictionary key named class_ids. We return that dictionary through the predictions parameter of tf.estimator.EstimatorSpec. The caller can then retrieve the prediction by examining the dictionary passed back to the Estimator's predict method.
EVAL
When model_fn is called with mode == ModeKeys.EVAL, the model function must evaluate the model, returning loss and possibly one or more metrics.
We can calculate loss by calling tf.losses.sparse_softmax_cross_entropy. Here's the complete code:
# To calculate the loss, we need to convert our labels
# Our input labels have shape: [batch_size, 1]
labels = tf.squeeze(labels, 1) # Convert to shape [batch_size]
loss = tf.losses.sparse_softmax_cross_entropy(labels=labels, logits=logits)
Now let's turn our attention to metrics. Although returning metrics is optional, most custom Estimators return at least one metric. TensorFlow provides a Metrics API (tf.metrics) to calculate different kinds of metrics. For brevity's sake, we'll only return accuracy. The tf.metrics.accuracycompares our predictions against the "true labels", that is, against the labels provided by the input function. The tf.metrics.accuracyfunction requires the labels and predictions to have the same shape (which we did earlier). Here's the call to tf.metrics.accuracy:
# Calculate the accuracy between the true labels, and our predictions
accuracy = tf.metrics.accuracy(labels, predictions['class_ids'])
When the model is called with mode == ModeKeys.EVAL, the model function returns a tf.estimator.EstimatorSpec containing the following information:
- the
mode, which is tf.estimator.ModeKeys.EVAL - the model's loss
- typically, one or more metrics encased in a dictionary.
So, we'll create a dictionary containing our sole metric (my_accuracy). If we had calculated other metrics, we would have added them as additional key/value pairs to that same dictionary. Then, we'll pass that dictionary in the eval_metric_ops argument of tf.estimator.EstimatorSpec. Here's the block:
# Return our loss (which is used to evaluate our model)
# Set the TensorBoard scalar my_accurace to the accuracy
# Obs: This function only sets value during mode == ModeKeys.EVAL
# To set values during training, see tf.summary.scalar
if mode == tf.estimator.ModeKeys.EVAL:
return tf.estimator.EstimatorSpec(
mode,
loss=loss,
eval_metric_ops={'my_accuracy': accuracy})
TRAIN
When model_fn is called with mode == ModeKeys.TRAIN, the model function must train the model.
We must first instantiate an optimizer object. We picked Adagrad (tf.train.AdagradOptimizer) in the following code block only because we're mimicking the DNNClassifier, which also uses Adagrad. The tf.trainpackage provides many other optimizers—feel free to experiment with them.
Next, we train the model by establishing an objective on the optimizer, which is simply to minimize its loss. To establish that objective, we call the minimizemethod.
In the code below, the optional global_step argument specifies the variable that TensorFlow uses to count the number of batches that have been processed. Setting global_step to tf.train.get_global_stepwill work beautifully. Also, we are calling tf.summary.scalarto report my_accuracy to TensorBoard during training. For both of these notes, please see the section on TensorBoard below for further explanation.
optimizer = tf.train.AdagradOptimizer(0.05)
train_op = optimizer.minimize(
loss,
global_step=tf.train.get_global_step())
# Set the TensorBoard scalar my_accuracy to the accuracy
tf.summary.scalar('my_accuracy', accuracy[1])
When the model is called with mode == ModeKeys.TRAIN, the model function must return a tf.estimator.EstimatorSpeccontaining the following information:
- the mode, which is
tf.estimator.ModeKeys.TRAIN - the loss
- the result of the training op
Here's the code:
# Return training operations: loss and train_op
return tf.estimator.EstimatorSpec(
mode,
loss=loss,
train_op=train_op)
Our model function is now complete!
The custom Estimator
After creating your new custom Estimator, you'll want to take it for a ride. Start by
instantiating the custom Estimator through the Estimatorbase class as follows:
classifier = tf.estimator.Estimator(
model_fn=my_model_fn,
model_dir=PATH) # Path to where checkpoints etc are stored
The rest of the code to train, evaluate, and predict using our estimator is the same as for the pre-made DNNClassifierdescribed in Part 1. For example, the following line triggers training the model:
classifier.train(
input_fn=lambda: my_input_fn(FILE_TRAIN, repeat_count=500, shuffle_count=256))
TensorBoard
As in Part 1, we can view some training results in TensorBoard. To see this reporting, start TensorBoard from your command-line as follows:
# Replace PATH with the actual path passed as model_dir
tensorboard --logdir=PATH
Then browse to the following URL:
localhost:6006
All the pre-made Estimators automatically log a lot of information to TensorBoard. With custom Estimators, however, TensorBoard only provides one default log (a graph of loss) plus the information we explicitly tell TensorBoard to log. Therefore, TensorBoard generates the following from our custom Estimator:
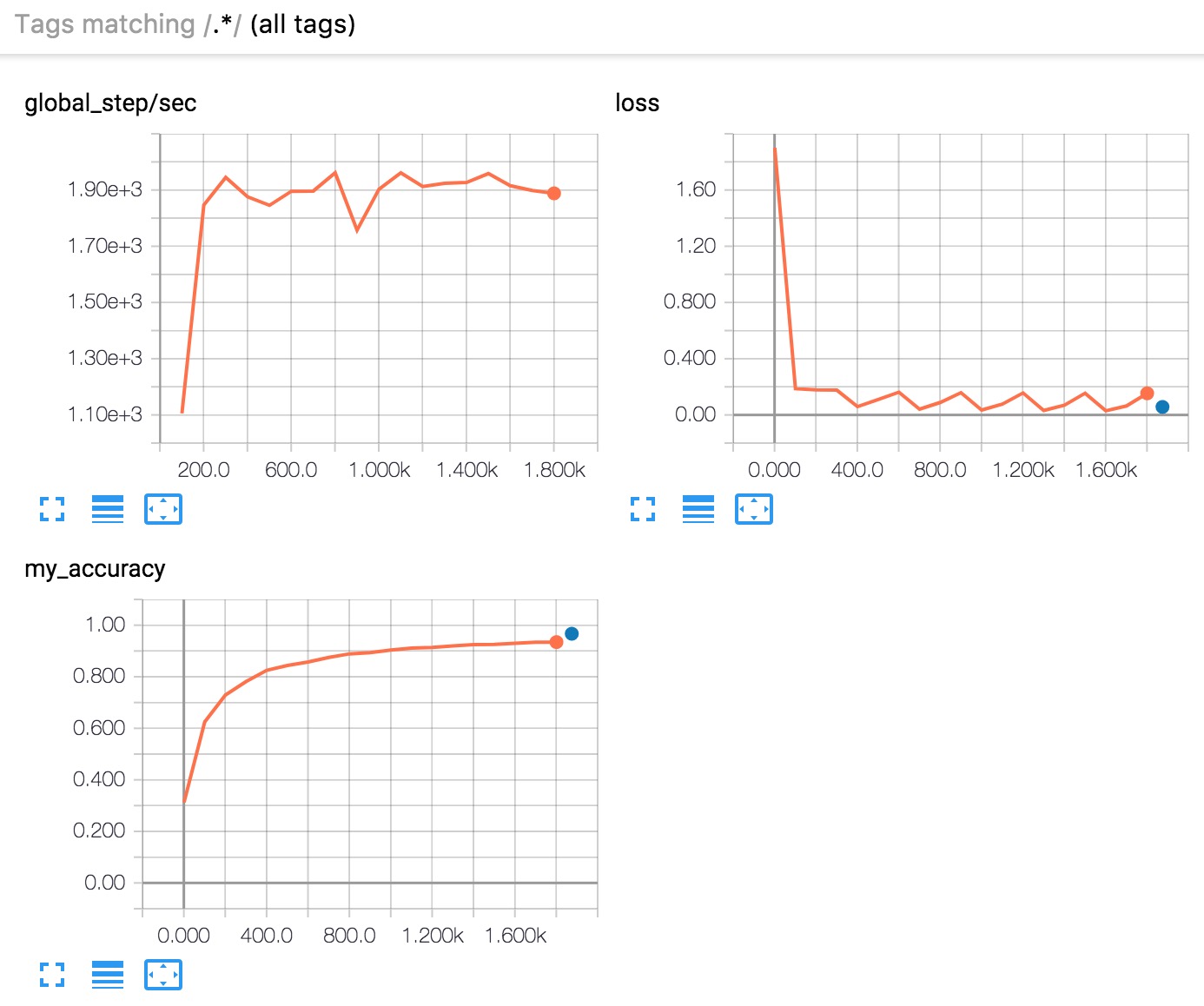
Figure 6. TensorBoard displays three graphs.
In brief, here's what the three graphs tell you:
- global_step/sec: A performance indicator, showing how many batches (gradient updates) we processed per second (y-axis) at a particular batch (x-axis). In order to see this report, you need to provide a
global_step (as we did with tf.train.get_global_step()). You also need to run training for a sufficiently long time, which we do by asking the Estimator train for 500 epochs when we call its train method: - loss: The loss reported. The actual loss value (y-axis) doesn't mean much. The shape of the graph is what's important.
- my_accuracy: The accuracy recorded when we invoked both of the following:
eval_metric_ops={'my_accuracy': accuracy}), during EVAL (when returning our EstimatorSpec) tf.summary.scalar('my_accuracy', accuracy[1]), during TRAIN
Note the following in the my_accuracy and loss graphs:
- The orange line represents
TRAIN. - The blue dot represents
EVAL.
During TRAIN, orange values are recorded continuously as batches are processed, which is why it becomes a graph spanning x-axis range. By contrast, EVAL produces only a single value from processing all the evaluation steps.
As suggested in Figure 7, you may see and also selectively disable/enable the reporting for training and evaluation the left side. (Figure 7 shows that we kept reporting on for both:)
Figure 7. Enable or disable reporting.
In order to see the orange graph, you must specify a global step. This, in combination with getting global_steps/sec reported, makes it a best practice to always register a global step by passing tf.train.get_global_step()as an argument to the optimizer.minimize call.
Summary
Although pre-made Estimators can be an effective way to quickly create new models, you will often need the additional flexibility that custom Estimators provide. Fortunately, pre-made and custom Estimators follow the same programming model. The only practical difference is that you must write a model function for custom Estimators. Everything else is the same!
For more details, be sure to check out:
Until next time - Happy TensorFlow coding!